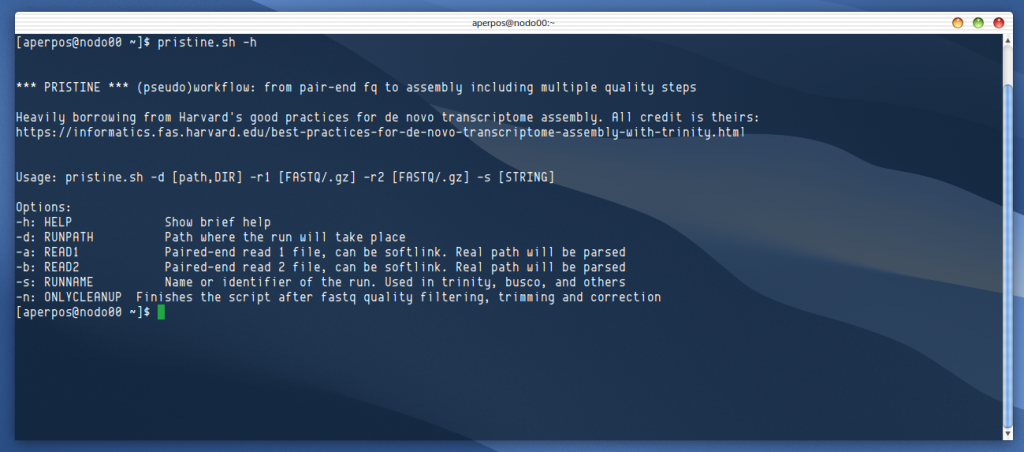
The scientific career seems to follow a pattern of slight variations, from the feeling of senseless wandering (at first overwhelmed by a sea of knowledge as an undergrad or phd student, but later as skepticism and solipsism based on first-hand experience), to the uncertainty of what comes after our current stage (postdoc? Young PI? Faculty?). As many say, it doesn’t get easier -but it can get better and more rewarding. And to spark a bit of hope in the steps following the completion of the PhD, today I am drawing from my experience of applying to five postdoctoral fellowships in the past few months of 2021, to summarise what I have learned, what can be improved, and whatever might be of help.
Continue reading