In my most recent publication, together with some formidable colleagues from my previous lab, we describe the origins of animals by compiling the state-of-the-art of current knowledge and methods. What is special about our work is that we do so by reconstructing two ancestors key to understand this evolutionary transition: the last unicellular ancestor of animals and the last common ancestor of all animals. In this entry, I have compiled some notes and paragraphs that were left out or simplified, but that were very informative and illustrative for me during the reading process of describing the last common ancestor of animals (animal LCA). They have been cohesively edited together adding some notions of evolutionary biology and ecology to provide a solid piece of read. Some parts are somewhat similar to what was published, in the end, this is a sub-field of what we did. I did my best to reflow and change those parts to avoid redundancy. I hope you enjoy it as much as I did!
0. Preamble on the origin animals and their last common ancestor
The origin of animals has long been discussed, and many factors have been proposed to explain why and how it occurred. The most cited arguments, such as an arms race for bigger sizes and expanding to new ecological niches, involve selection-oriented mechanisms, that is, the classical forces of evolution.
The origin of animals involved the consolidation of multicellularity, a major evolutionary transition from single-celled organisms (for the most part of their life cycle) to multicellular organisms with a complex developmental plan and spatial co-occurrence of cell types. This increase at the organizational level entailed the emergence of a new evolutionary space at the level of whole groups of cells. In other words, selection started operating in new aspects related to the consequences of the actions of multiple cells together, rather than the outcome of the activity of single cells. This leads to the emergence of selection between groups.
Thanks to our increasing knowledge of the genomes of animals and their unicellular relatives, we now know the transition was at least partially driven by a complexification of genome features and genome architecture. Because this complexity is an a-priori situation, we cannot invoke the aforementioned Darwinian properties as means to explain how it originated (Clarke, 2014; Griesemer, 2001; Libby and Rainey, 2011; Rainey, 2007; Rainey and Kerr, 2010). The challenge of finding the mechanisms that led to this complexity can be alleviated by arguing a most plausible case of neural constructive evolution, whereby different elements of a network (such as genes and their regulatory elements) arise in complexity through non-adaptive steps (Muñoz-Gomez et al., 2021). The ancestors of animals comprised a stem lineage with pre-adaptive value as their regulatory program had the potential for their parts, originally non-dependent, to become increasingly interactive throughout non-adaptive unidirectional changes (Gray et al., 2010, Lukes et al., 2011).
In summary, the evolutionary history of animals showcases a number of drivers different in nature intertwined at multiple scales. How can we better understand the interplay between these evolutionary forces at the genetic, cellular, and organismal level? One possibility is by reconstructing their last common ancestor, which we call the animal LCA (see Box 1 in the Review). Such reconstruction allows to pinpoint the specifics of what features were crucial in the rise of animals and when they emerged. In turn, this helps to 1) build the minimal set of features that led to the radiation in diversity we see today in animals, and 2) shed light into the phylogenetic relationships of the earliest-branching animal lineages.
In this entry I focus on the reconstruction of the last common ancestor of animals based on the latest news from genomics and single cell studies. I highlight/emphasize some details regarding the main genetic aspects that make multicellular animal life (cell adhesion, signaling, communication, development, genomic regulation) and wrap up commenting on the connections in our latest work alongside some ecological aspects.
1. What do we know at the historical level?
All animals living today share a common multicellular ancestor, that we denominate the animal LCA, and that followed the consolidation of multicellularity from a unicellular ancestor. This transition occurred more than 600 million years ago (Ma), although there is still no exact consensus between the fossil record and biochemical and molecular clock-based estimates.
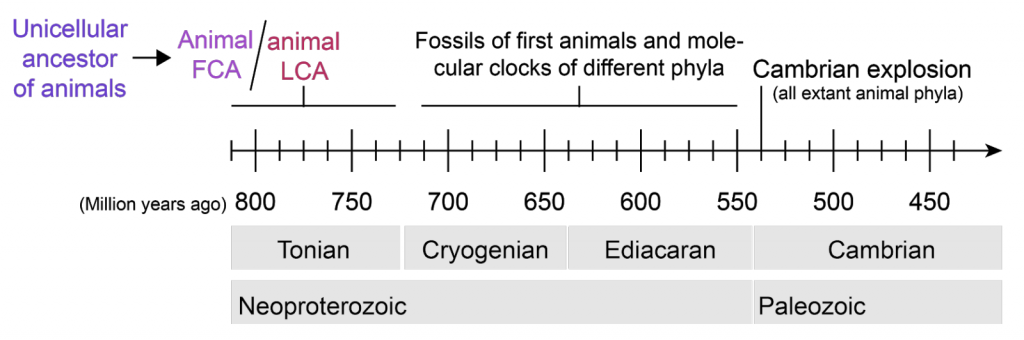
Until recently, fossil record inferences were the only way to address when the animal LCA originated and what it looked like. The first unequivocal evidence of animals in the fossil record date back to the Ediacaran period, with molecular clocks extending the estimate of the emergence of animals by more than a 100 million years, during the Cryogenian or the Tonian periods (Budd and Jensen, 2017; dos Reis et al., 2015; Douzery et al., 2004; Erwin et al., 2011; Knoll, 2011; Lozano-Fernandez et al., 2017; Knoll and Hewitt, 2013; Narbonne, 2005; Peterson et al., 2004; but see e.g., Eme et al., 2014 for a discussion of the caveats in molecular-clock-based estimates). Some examples of these animal macrofossils are Dickinsonia, Kimberella (both from around 560 Ma), Eoandromeda octobrachiata (~555 Ma) or Eocyathispongia qiania (~600 Ma), whose body plans suggest that axial patterning and symmetry were featured early in animal evolution (Fedonkin and Waggoner, 1997; Dunn et al., 2018; Ou et al., 2015; Tang et al., 2011; Yin et al., 2015).

Although we cannot work with the living bodies of extinct organisms, there are molecules that can survive the passing of such long time, and their evidences can still be found in their fossils. Although the use of sterols as biomarkers of animal life in the fossil record remains controversial (Nettersheim et al., 2019), molecular characterization of the sterol composition of ancient sponge-like fossils and Dickinsonia fossils suggests that these organisms have animal sterols and therefore share ancestry with animals living today (Bobrovskiy et al., 2018; Love et al., 2009). Thus, all extant animal lineages share a common ancestor that lived on Earth prior to the Cambrian explosion, ca. 700 Ma, greatly diversifying later during the Ediacaran and Cambrian period (Budd and Jensen, 2017; Narbonne, 2005).
2. What do we know at the genetic level?
Despite the limited information provided by the fossil record about the biology of the first animals, we can reconstruct part of it by looking at the extant, earliest branching animals living today. All animals have a body plan relying on symmetry at least during a life stage, which allows a clear distinction between non-bilaterians and bilaterians. Under this scenario, and before confirmation by molecular phylogeny, animals with more rudimentary body plans (that is, sponges, ctenophores, cnidarians, and placozoans) were considered the earliest branching lineages. Studies in sponges prompted authors like Muller (Muller, 2004) to speculate about the nature of the first animals. Early discussions in the field pointed that evolution of cell-cell and cell-matrix adhesion mechanisms was needed prior to the emergence of animals. Likewise, evolution of kin recognition and immunity through cell signaling was also crucial for the emergence of biological entities at the multicellular level. Stepwise acquisition of gene regulation involving these processes might have lead to an organism with the potential to radiate in the diversity we see today, both in extant lineages and in the fossil record.
To uncover the nature of the animal LCA beyond inferences from fossils, classical approaches have focused on comparing the earliest branching animal lineages (including sponges, ctenophores, placozoans and cnidarians) with bilaterian animals. Advances in comparative genomics have provided deeper insight into the molecular characteristics of these lineages, with dozens of non-bilaterian genomes and transcriptomes currently available (e.g., Kamm et al., 2018; Kenny et al., 2020; Chapman et al., 2010; Moroz et al., 2014; Putnam et al., 2007; Riesgo et al., 2014; Ryan et al., 2013; Srivastava et al., 2010, 2008; Gold et al., 2019; Leclère et al., 2019). As pointed by (Dohrmann and Wörheide, 2013), understanding the relationships of the metazoan basal lineages helps to understand how metazoan traits arose and evolved during evolution. However, a lack of consensus on the evolutionary relationships between some of these lineages (Telford et al., 2015) has somewhat hindered the reconstruction of metazoan trait emergence and evolution (Dohrmann and Wörheide, 2017). For instance, the debate over the earliest split in the metazoan tree of life is long and still ongoing – either Ctenophora or Porifera are the sister group of the rest of Metazoa (Laumer et al., 2019; Moroz et al., 2014; Pisani et al., 2015; Ryan et al., 2013; Telford et al., 2015; Whelan et al., 2015; King and Rokas, 2017). Despite remaining unresolved, reconstruction of the animal LCA is still possible in many other aspects (King and Rokas, 2017) regardless of the scenario considered.
The genome sequencing of several animal species, together with the discovery and sequencing of animals’ closest unicellular relatives (see in the Review), has contributed significantly to reconstructing the macroevolutionary mechanisms orchestrating animal genome evolution and the animal LCA gene toolkit.
In the next post, we will go through a detailed overview of what have we learned from animal genomes, both in a broad sense of what changes in the genome entailed the evolution of early animals, and what were the minimum tools in the gene kit of the last common ancestor of animals.


Thank you!